Zong-Yan Liu.
I build things for the plants
A dedicated PhD candidate at Cornell University, I leverage machine learning techniques to unravel the complexities of plant genomes and their evolutionary history.
A dedicated PhD candidate at Cornell University, I leverage machine learning techniques to unravel the complexities of plant genomes and their evolutionary history.


I am a fourth-year Ph.D. candidate at Cornell University in the Buckler Lab with a passion for computational biology and genetics. My background bridges computer science and plant genetics, and I’m deeply interested in how data science can advance our understanding of crop genomes. Currently, I am conducting research in maize genetics, focusing on genomic prediction and statistical modeling to improve crop resilience.
I am working on a project that utilizes advanced computational techniques to enhance genomic analysis in maize, aiming to contribute to sustainable agriculture and food security.
Here are a few technologies I've been working with recently:I serve as the President of Synapsis, where I lead initiatives to build a supportive community and enhance the professional and academic experiences of graduate students in Cornell Plant Breeding and integrative plant sciences.
I serve as the President of the Cornell Taiwanese Student Association (CTSA), where I lead initiatives to foster community and cultural exchange, organize events and resources to support Taiwanese and international students, and advocate for their needs and interests within the Cornell University environment.
I am a Graduate Research Assistant at Academia Sinica’s Institute of Information Science, where I focus on studying extrachromosomal circular DNA (eccDNA) for cancer prediction. Additionally, I am responsible for maintaining our lab’s database, ensuring data integrity and accessibility to support ongoing research initiatives.
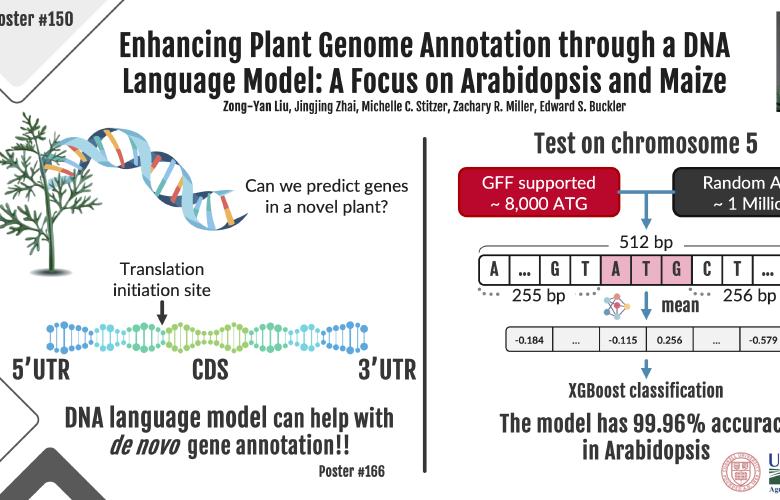
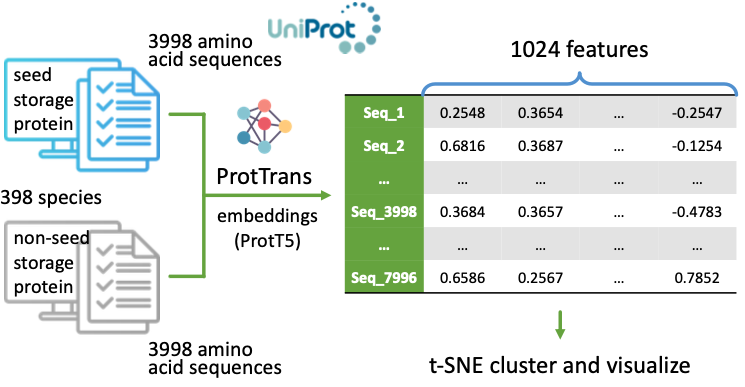
Focuses on developing advanced computational models to accurately predict gene structures and protein abundance in plants, enhancing our understanding of plant genomics and providing valuable insights into crop biology and evolution.
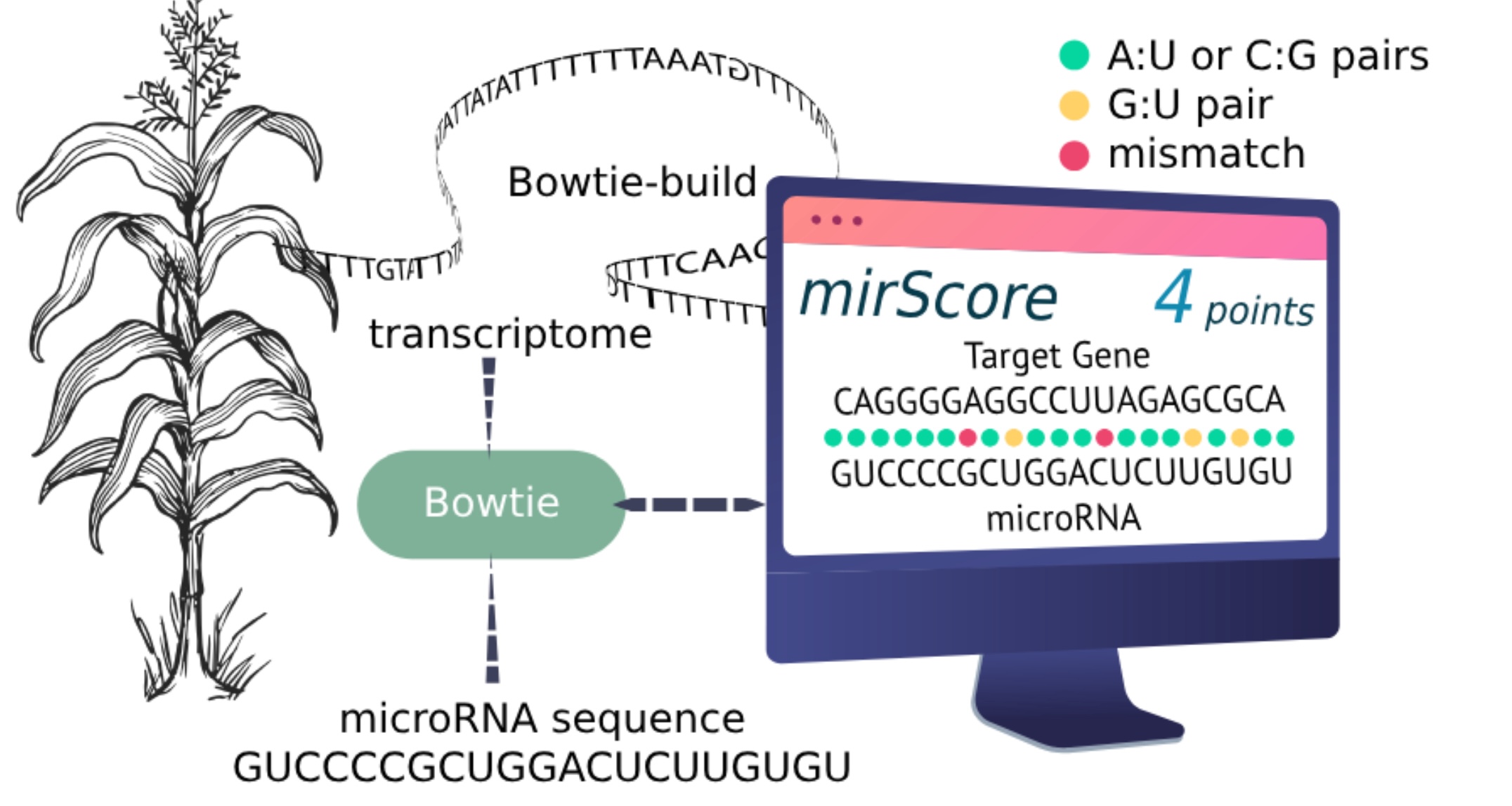
Analysis of IbHypSys-mediated MicroRNAs upon Wounding in Sweet Potato (Ipomoea batatas cv. Tainung 57)
Majoring in Agronomy with a double minor in Management Information Systems and Applied Economics.
Research in Agronomy