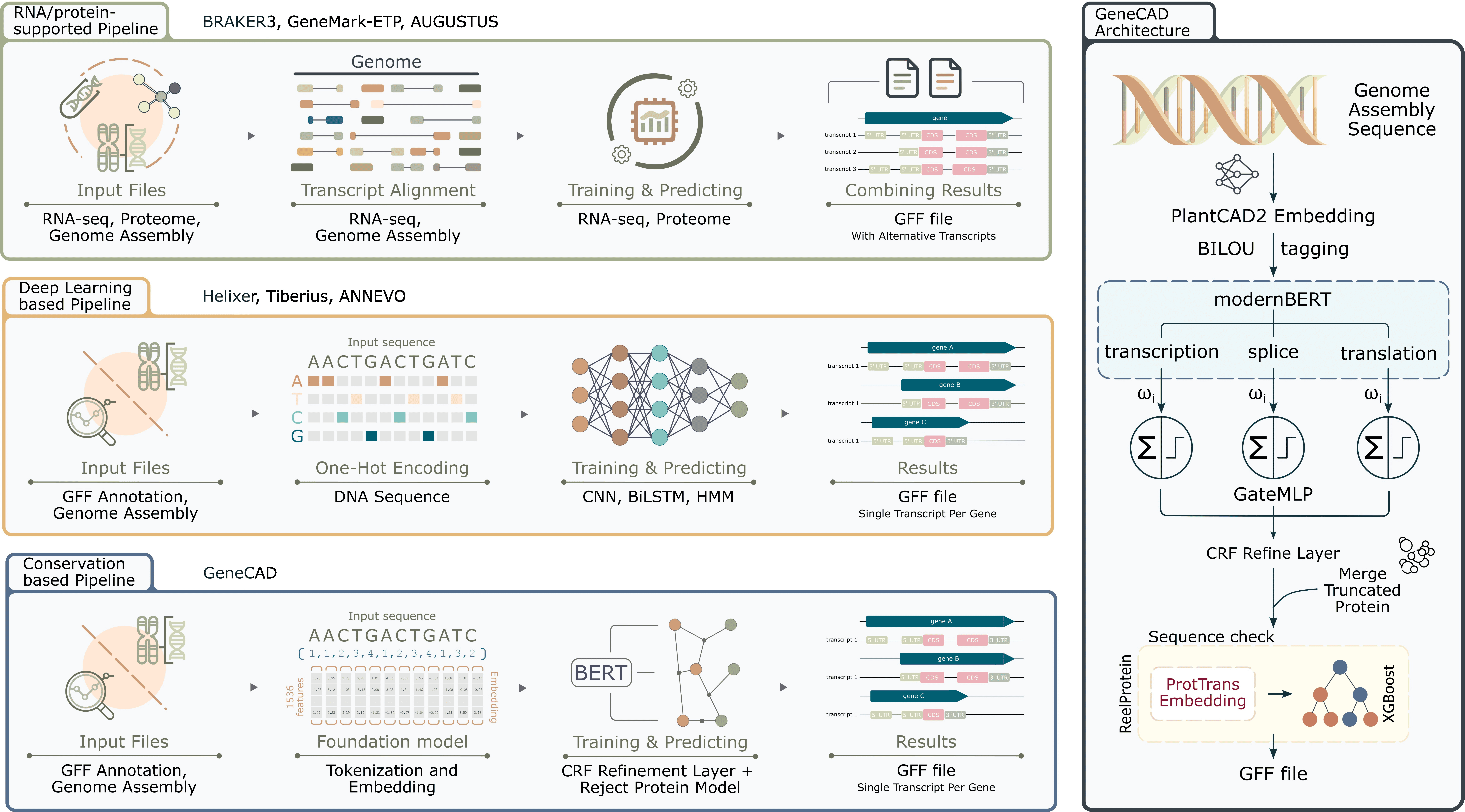
GeneCAD at a glance
GeneCAD is a sequence-only annotation pipeline that turns genome sequence into complete, biologically coherent plant gene models (GFF3) using a DNA foundation model (PlantCAD2), a ModernBERT head, and a chromosome-wide CRF—no RNA-seq, no proteomics, no homology inputs needed.
- What’s new: the first foundation-model / LLM–based workflow that assembles full plant gene models directly from DNA sequence and outperforms state-of-the-art baselines in cross-species tests.
- Data efficiency: trains effectively on a small, curated set of high-quality references using a masked-motif logistic regression (MMLR) score to reduce label noise.
- Scale & robustness: accurate across diploid and polyploid genomes; boundaries are sharp at start/stop codons and splice junctions.
Preprint: https://doi.org/10.1101/2025.10.31.685877
Code: https://github.com/plantcad/genecad
How GeneCAD works
- Representation: Plant genomes are embedded with PlantCAD2 to capture conservation-aware sequence signals across angiosperms.
- Head & labels: An 8-layer ModernBERT predicts BILOU tags for CDS, introns, and 5′/3′ UTRs at single-nucleotide resolution.
- Structure-aware decoding: A chromosome-wide CRF enforces splice-phase continuity and legal feature order to yield coherent transcripts.
- Protein plausibility screen: Predicted CDSs are filtered by a protein language-model plausibility score to suppress repeat-driven ORFs and boost precision.
Why it’s a breakthrough
- DNA-only input: Accurate genome annotation without matched RNA-seq or proteomics—ideal for scaling across species with scarce assays.
- Quality over quantity: MMLR-curated training data delivers strong performance without “tons of data.”
- Cross-species generalization: Maintains accuracy across diverse clades and ploidy while keeping boundaries precise.
Benchmarks (held-out angiosperms)
Across five held-out species (including the allotetraploid Nicotiana tabacum), GeneCAD improves transcript-level F1 over Helixer and BRAKER3, increases exact-match transcripts, and sharpens start/stop and splice boundaries. Even when training is reduced from five curated genomes to two species, most accuracy is retained.
Collaboration & acknowledgments
This work is a collaboration between Cornell University / Institute for Genomic Diversity and the Open Athena AI Foundation, with thanks to partners who supported compute and community deployment.
How to cite GeneCAD
Liu, Z.-Y., Berthel, A., Czech, E., Stitzer, M. C., Hsu, S.-K., Pennell, M., Buckler, E. S., & Zhai, J. (2025). GeneCAD: Plant Genome Annotation with a DNA Foundation Model. bioRxiv. https://doi.org/10.1101/2025.10.31.685877