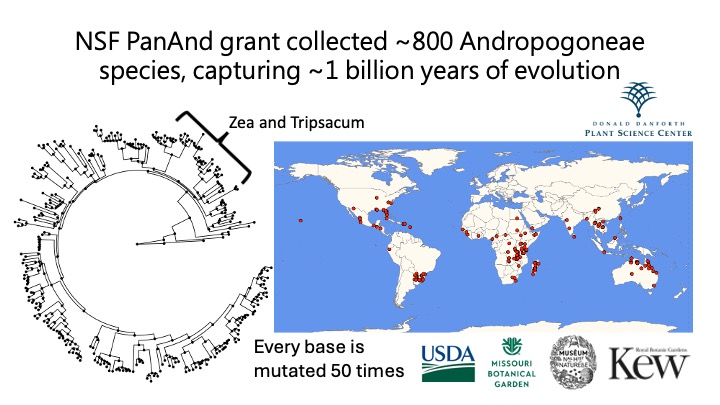
Overview
Our latest preprint, “Extensive genome evolution distinguishes maize within a stable tribe of grasses,” presents a comprehensive analysis of 33 genome assemblies from 27 species within the Andropogoneae tribe. The study captures a dynamic picture of genome evolution, revealing how maize (and its close relatives) navigates the challenges and surprises of polyploidy.
Key Highlights
Polyploidy Unraveled:
The study examines 14 independent polyploid formation events, comparing diploids, tetraploids, hexaploids, and paleotetraploids. Contrary to traditional expectations, most polyploid events exhibit a remarkable stability in chromosome number, gene retention, and overall genome organization.Chromosome Dynamics:
- Stability over Reduction: While some genomes undergo chromosomal fusions, the majority retain multiples of the base chromosome number (x=10).
- Rearrangements: In-depth analyses show that the maize lineage experienced massive rearrangements, such as the fusion of entire ancestral chromosomes, setting it apart from other members of the tribe.
Gene and Repeat Content:
- Despite polyploidy, gene loss is not as extensive as expected; many duplicate genes are retained, especially those involved in key developmental and regulatory processes.
- Transposable Elements (TEs): The study finds that TE expansion is not a universal consequence of polyploidy. In fact, only select lineages show bursts of TE activity, while others accumulate TE sequences gradually over time.
Regulatory Sequences in Flux:
Noncoding regions, including transcription factor binding sites (TFBS), turn over rapidly compared to coding sequences. This suggests that while gene content remains relatively stable, the regulatory landscape is highly dynamic and may drive adaptive changes.
Major Findings
Polyploid Evolution is Not One-Size-Fits-All:
- Unexpected Stability: Most polyploid Andropogoneae species do not follow the anticipated patterns of rapid gene loss or chromosomal reduction.
- Diverse Outcomes: In contrast to the maize lineage, several polyploid events exhibit minimal large-scale genomic reorganization.
Chromosome Rearrangement Patterns:
- Maize exhibits a drastically rearranged karyotype with a reduced haploid chromosome number, highlighting the role of chromosomal knobs and neocentromeres in mediating genomic changes.
- Other polyploid species show a more conservative approach, retaining a high number of chromosomes with fewer rearrangements per chromosome.
TE Accumulation and Regulatory Changes:
- Although polyploidy can provide a buffering effect against deleterious TE insertions, the rate and timing of TE amplification vary widely.
- The rapid turnover of regulatory elements, especially TFBS, points to an evolutionary flexibility that may underlie maize’s remarkable adaptability.
Impact on Plant Genomics
This research challenges the classic models of polyploid evolution by showing that:
- Genomic stability can coexist with polyploid complexity.
- The evolutionary fate of duplicate genes and regulatory sequences is influenced by a combination of chromosomal, epigenetic, and environmental factors.
- Understanding these dynamics provides valuable insights into crop evolution, adaptation, and ultimately, breeding strategies for enhanced resilience and productivity.
Data & Methods Snapshot
The study combines:
- High-quality genome assemblies: 33 assemblies representing 27 species.
- Cutting-edge sequencing and scaffolding techniques: PacBio HiFi, CLR, and Oxford Nanopore sequencing, along with BioNano optical mapping and Hi-C scaffolding.
- Robust bioinformatics pipelines: Gene annotation via deep learning models, synteny analyses using AnchorWave, and advanced TE and regulatory element characterization.
Note: Read the Full Preprint For those interested in delving deeper, access the full preprint here:
https://www.biorxiv.org/content/10.1101/2025.01.22.633974v1.full