Revolutionizing Cross-Species Plant Genomics 🌿
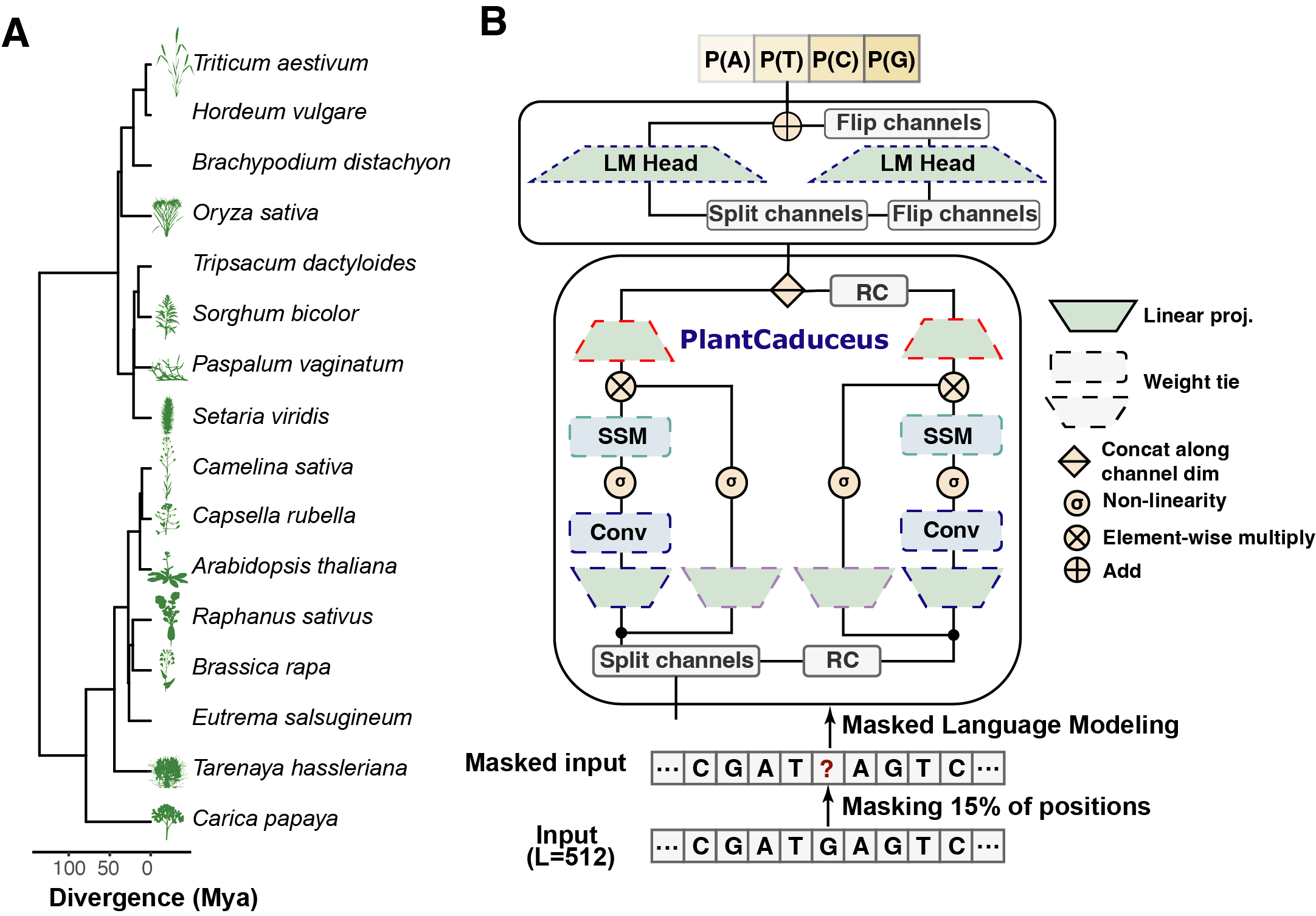
PlantCaduceus is an advanced platform designed to model plant genomes across species at single nucleotide resolution. By leveraging pre-trained DNA language models, PlantCaduceus aims to capture the evolutionary conservation of plant genomes, enabling powerful cross-species insights. This approach significantly accelerates the identification of functional genomic elements, ultimately supporting the fields of agriculture, plant breeding, and genomic research.
Cross-Species Modeling at Its Finest
PlantCaduceus builds upon Caduceus and Mamba architectures to analyze 16 Angiosperm genomes, covering evolutionary histories spanning 160 million years. This extensive dataset allows PlantCaduceus to understand genome complexities and identify crucial genetic variations. With tools like masked language modeling, PlantCaduceus offers a robust way to improve genome annotations and make valuable biological predictions, helping researchers pinpoint causal mutations, as in the case of the sweet corn mutation.
Pioneering Zero-Shot Genomic Analysis
By integrating a zero-shot learning approach, PlantCaduceus predicts deleterious mutations without needing specific training datasets for each plant species. This innovative method shows a three-fold enrichment of rare alleles compared to traditional approaches, demonstrating PlantCaduceus’s effectiveness in prioritizing important genetic mutations. This potential makes PlantCaduceus an invaluable resource for cross-species genomic research, bringing novel insights to evolutionary biology and improving plant resilience in agriculture.
For more details, visit PlantCaduceus.
How to cite PlantCaduceus
Zhai, J., Gokaslan, A., Schiff, Y., Berthel, A., Liu, Z. Y., Lai, W. L., Miller, Z. R., Scheben, A., Stitzer, M. C., Romay, M. C., Buckler, E. S., & Kuleshov, V. (2025). Cross-species modeling of plant genomes at single nucleotide resolution using a pretrained DNA language model. Proceedings of the National Academy of Sciences, 122(24), e2421738122. https://doi.org/10.1073/pnas.2421738122