

Hi, thank you for taking a look around my site. If you have any questions feel free to reach out on LinkedIn, Github, or by email!
New website
Enhancing Plant Genome Annotation through DNA Language Model: A Focus on Arabidopsis and Maize
Extrachromosomal Circular in Human Cancer Prediction
Analysis of IbHypSys-mediated MicroRNAs upon Wounding in Sweet Potato (Ipomoea batatas cv. Tainung 57)
0
Publication
0
Leadership
0
Project
0
Award
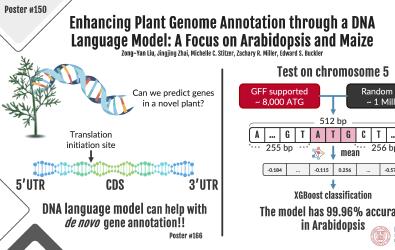
Advancements in genome sequencing have greatly facilitated the study of organisms, yet understanding genomic variation remains complex, particularly in genome annotation which models genetic transcription and translation. We refined a machine learning model, the Genomic Pre-trained Network (GPN), improving gene annotation in plant genomes by utilizing Ribosome profiling data to accurately identify gene initiation and stop sites, demonstrating a 92% prediction accuracy.
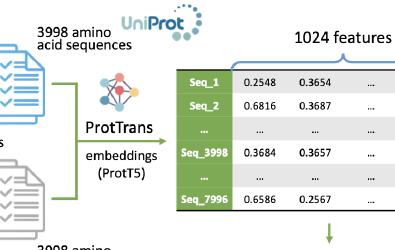
Seed storage proteins, crucial for plant development and as a food source, are abundantly found in crops like wheat and maize. However, their diversity in different plant species is not fully understood. We used the ProtTrans tool and a support vector machine classifier to analyze their physicochemical properties, aiming to identify new storage proteins in the UniProt database and Andropogoneae genomes, enhancing our knowledge of these essential proteins.
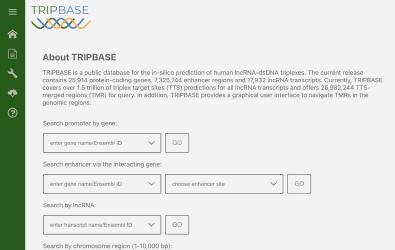
Long-non-coding RNAs (lncRNAs) are crucial for various biological processes and can form triple helices with DNA, but computational predictions often result in high false positives. By analyzing experimental data with Triplexator and applying six new filters, we improved the accuracy of these predictions. Consequently, we developed TRIPBASE, a database for exploring human lncRNA triplex formations with customizable filtering options, enhancing research into genomic cis-regulatory regions.